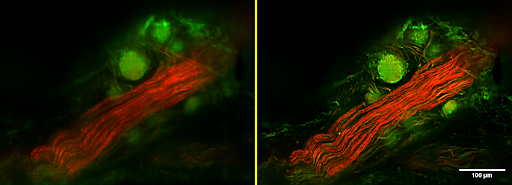
Supporting faster paths to heart disease insights
The below case study shares some of the technical details and outcomes of the scientific and HPC-focused programming support provided to a research project through NeSI’s Consultancy Service.
This service supports projects across a range of domains, with an aim to lift researchers’ productivity, efficiency, and skills in research computing. If you are interested to learn more or apply for Consultancy support, visit our Consultancy Service page.
Research background
University of Auckland researcher Greg Sands studies the structure of heart tissues and diseases. His goal is to understand structural changes that occur in heart disease and which affect the electrical function of the heart.
With his colleagues at the Auckland Bioengineering Institute, Greg developed a novel scanned-line confocal microscope that enables high-resolution 3D imaging at multiple wavelengths over scales exceeding those available using standard confocal or light-sheet systems. Image resolution is typically better than 1 µm, and images can span tens of millimetres in-plane and several millimetres axially, with image sizes often approaching one trillion 3D pixels. The same imaging technology is now also being applied to brain, lymph, and other tissues to look at their own structure-function relationships.
Project challenges
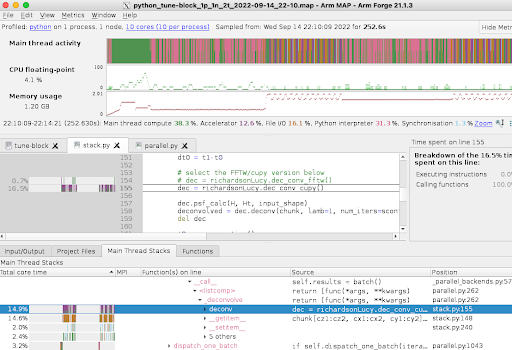
Greg and colleagues have developed a parallel Python code which reconstructs images from the raw data, including deconvolution using the microscope point spread function. Whilst the code scales very well, the sheer size of the images (100 GB to TBs) means that there is a backlog of images to be processed. Small cases run with 60 processors require about 30 hours to complete, while larger images take 10x longer. The picture below shows the output of the ARM MAP profiler, highlighting the execution bottlenecks.
What was done
NeSI Research Software Engineer Alexander Pletzer and Data Science Engineer Maxime Rio were tasked to evaluate the present implementation of the code and explore whether additional performance improvements could be obtained. They focused their attention on the Fast Fourier Transform (FFT) parts of the algorithm, which applies a deconvolution kernel to sharpen the image. They found that, although the implementation was optimal for running on a Central Processing Unit (CPU), migrating the code to a Graphical Processing Unit (GPU) could accelerate the code.
Main outcomes
Following this Consultancy project, the researchers can now run their code on either CPU or GPU resources. On the GPU, the code runs 3.5x faster than on the CPU. Having a choice between running on CPUs or GPUs will enable the researchers to optimise their use of supercomputer resources, depending on what type of resource is available and their needs at the time. Ultimately, having the ability to process microscope images faster and in larger quantities improves researchers' ability to deliver benefits to patients and contribute insights to other research projects in this field.
Researcher feedback
"The consultation process was valuable in many ways. Simply to have an outside perspective on our problem and code enabled significant progress, not only in the execution time, but in the quality, efficiency and robustness of the computation. The consultants quickly grasped the scope of our problem and identified alternative approaches that were unfamiliar to us. Their engagement prompted us to review our coding and testing methods which has an impact beyond just this project. We are now able to maximise our use of the NeSI platform, and run computations on a scale that was not previously possible."
- Greg Sands, Senior Research Fellow, Auckland Bioengineering Institute, University of Auckland
Do you have a research project that could benefit from working with NeSI Research Software Engineers or our Data Science Engineer? Learn more about what kind of support they can offer and get in touch by emailing support@nesi.org.nz.