How Genomics Aotearoa and NeSI tackled a big problem (and won)
The Challenge:
Assemble the first stick insect genome using linked-read technology in a way that is accurate, efficient, and reusable for other genomes.
NeSI's Solution:
Access to dedicated high-memory compute resources and bioinformatics expertise.
The Impact:
Genomics Aotearoa can now identify other genomes that could be sequenced and assembled using these genomic technologies, to contribute to conservation and selective breeding activities in primary industries.
A major goal for Genomics Aotearoa (GA) is to establish genomics analysis tools and pipelines that can be used by other research. Taking a “building by doing” approach, this means the tools, procedures and pipelines that are optimised and used for GA research projects are documented and made available for use in future projects.
NeSI’s collaboration with GA aims to support this strategy, so when GA Postdoctoral Researcher Ann McCartney approached NeSI with a request to use its computing platform to run a new de novo sequencing application (Chromium10X linked-read technology), the NeSI team saw an opportunity to deliver a solution with wide-reaching benefits.
At Manaaki Whenua - Landcare Research in Auckland, Ann is working alongside team leader Thomas Buckley to assemble a gold standard reference for the stick insect genome, as part of GA’s High Quality Genomes project.
This work is important for conservation and selective breeding within our primary industries because the higher quality the genomes, the greater the potential impact for research in conservation. The challenge, however, is that many of these genomes are large, very complex, repetitive, or highly variable which make them ‘difficult’ to sequence and assemble.
A supernova problem
Stick insects are biologically interesting. In times of stress, they have the ability to become parthenogenic, meaning the females lay eggs without needing to mate with males to produce offspring. They are also dynamic in a range of temperatures and altitudes in New Zealand. Better understanding of stick insect genomics will contribute much to global knowledge on the Phasmatodea species, including biogeographic origin, reproduction and temperature tolerance and its role in climate change.
Ann was planning assemblies using Chromium10X linked-read technology Supernova on four endemic New Zealand species (the hihi bird and three types of stick insects — Niveaphasma, Clitarchus, and Acanthoxyla). To do this, she needed additional long-read sequencing technology to ensure high quality genomes.
NeSI currently provides GA researchers with access to dedicated high-memory compute resources. However, the huge size of Ann’s stick insect genomes meant long-read would have been too ‘expensive’, or computationally demanding, whereas short read was not sufficient to provide the accuracy required. Linked-reads were chosen as a compromise between short-read and long-read sequencing technologies as they provide pseudo-long reads for a fraction of the price.
This still posed a challenge, however, as de novo assemblies that provide this option can be very demanding from an input/output (I/O) and memory perspective. They also often don’t fit well into traditional high performance computing modes of delivery, with characteristics such as very long run-times and difficult to predict resource requirements. Not only did Ann’s planned assemblies lack a reference genome, they were also some of most complex genomes to date, at a scale beyond even what the software vendor had supported so far.
Tackling the challenge
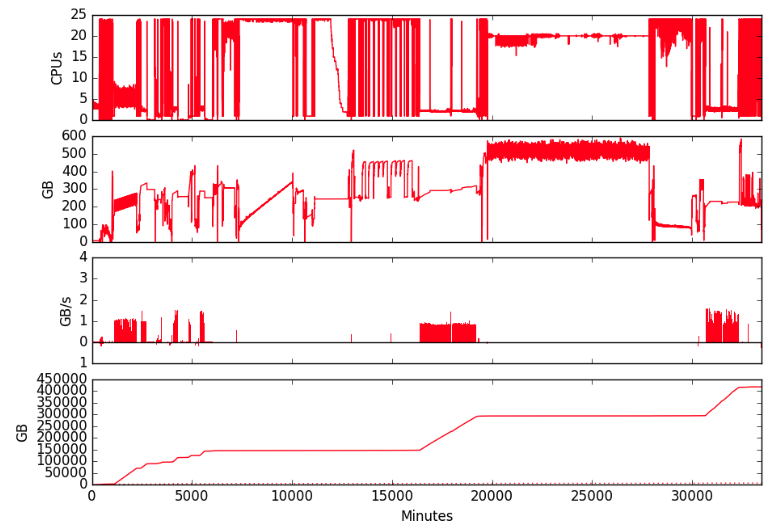
To start, NeSI threw a 4TB, 64-core node at the problem — NeSI calls these hugemem compute nodes. These nodes are a particularly important platform capability, enabling scientists to tackle big problems by scaling up rather than out.
There were several issues to sort after figuring out some of the nuances of how best to craft the parameters for each de novo assembly, taking memory and CPU requirements into account. One problem was that the application would stall part way through the pipeline, causing a whole node failure that forced a system reboot.
After several unsuccessful attempts tuning various aspects of the system, NeSI platform engineers identified they were hitting a system level deadlock likely due to a bug somewhere in the IBM Spectrum Scale filesystem (nee GPFS) client drivers. Addressing this required an upgrade of Spectrum Scale, something that could only be done during a complete outage of the platforms. Fortunately, NeSI had already scheduled maintenance work and so this upgrade was added to the list.
On Ann’s side, it took her nearly three months to adapt her techniques to get the first test of the genome assembly running.
“The computational environment required for these analyses is very complex, so we have had to research what is available, create environments and test under different parameters – there has been a lot of trial and error for this genome," she said.
As a result of her collaboration with the NeSI team, Ann’s stick insect genome assembly was successfully completed after 22 days running — the longest job to be run on the NeSI platform to date and the first stick insect genome assembly created using linked-read technology.
Despite having no prior experience with Supernova, the NeSI team’s combination of bioinformatics and platform resource expertise meant they could successfully address the barriers facing Ann’s project and provide a solution that will benefit other genomics researchers in New Zealand.
“Honestly, I have worked on a lot of clusters and I have never had the type of user support that NeSI supply,” she said. “This pipeline would not have been possible without the collaboration between myself and the NeSI support team and now it is possible to do anything Chromium10X assembly using the NeSI platform. Using NeSI has also significantly decreased the amount of computational power (RAM, storage, and CPU) that I need to have personally, as facilities such as NeSI can now carry out these types of computation.”
The implications of using linked-read technology
How is having this stick insect genome better than any other? The large and highly repetitive stick insect genome was the perfect test as to whether psuedo-long or linked-read technology were a more appropriate sequencing platform for genomes of this nature.
Due to its success, other endemic New Zealand species are now being sequenced using this technology, including the blueberry, the hihi, the myna, and the rewarewa.
- - - - - -
Do you have an example of how NeSI platforms or expertise have supported your work? We’re always looking for projects to feature as a case study. Get in touch by emailing support@nesi.org.nz.







