Applying machine learning to uncover molecular property insights
The below case study shares some of the technical details and outcomes of the scientific and HPC-focused programming support provided to a research project through NeSI’s Consultancy Service.
This service supports projects across a range of domains, with an aim to lift researchers’ productivity, efficiency, and skills in research computing. If you are interested to learn more or apply for Consultancy support, visit our Consultancy Service page.
Research background
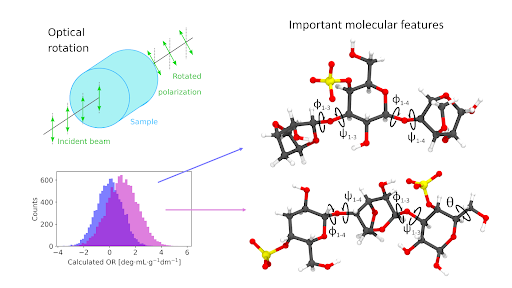
At Massey University, Dr. Benjamin Westberry is studying optical properties of carbohydrate polymers, in particular the polysaccharide κ-carrageenan. Carrageenan solutions are of particular interest in the food, pharmaceutical, and medical industries due to their tunable viscoelasticity. Studying a molecular property called optical rotation, the mechanisms at play behind the structure-function relationship can be elucidated. .
Computation of the optical properties of molecules can be achieved numerically using structures predicted via molecular dynamics simulations (MD) coupled to quantum mechanical (QM) calculations. Benjamin leveraged the NeSI high performance computing (HPC) platform to run these very intensive simulations. With this synthetic dataset, he then wants to understand the principle predictors of the optical rotation given the atomic structure of each molecular conformation. In this endeavour, Benjamin was keen on testing machine learning as a tool to help him analyse his dataset.
Project challenges
Leveraging machine learning (ML) as a tool to explain molecular properties imposes a set of methodological constraints. The ML models employed need to be interpretable or be compatible with explainable artificial intelligence (AI) techniques.
Since the dataset was small, the ML models would also need to be capable of operating in a small sample size regime and not be prone to overfitting (i.e. avoid learning the training data by heart, to be able to generalise to new data). Finally, and most importantly, ML models need to have good predictive performances to make the analysis of their properties meaningful.
Benjamin partnered with NeSI to overcome these challenges. With the help of NeSI Data Science Engineer Dr. Maxime Rio and NeSI Research Software Engineer Dr. Chris Scott, they designed and implemented an ML pipeline to evaluate a set of ML models in a systematic way and extract insights from the most promising models.
What was done
As a first step, Maxime implemented a reproducible ML pipeline using the Snakemake workflow management system. This pipeline consists of multiple steps to respectively prepare the dataset, split it into multiple sets for cross validation, fit models to each training set, evaluate models performances, and display the results.
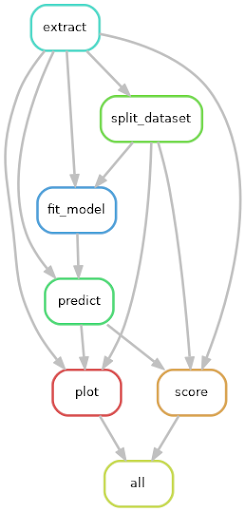
Many of these steps can be executed in parallel. Snakemake, via its support for the Slurm job scheduler, made it possible to distribute these computations on NeSI’s HPC platform, taking care of submitting jobs once their prerequisites were available.
Additionally, Snakemake tracks the state of generated files, ensuring that only steps whose input files are newer are recomputed. This made it very easy to incrementally add new models to the pipeline and avoid unnecessary recomputations.
In terms of models, Benjamin and Maxime settled on a set of standard interpretable models applied to features specific to molecular data. Tested models comprised elastic net linear regression models (EN), random forest models (RF), extremely randomised trees models (ERT), and multi-layer perceptron models with rectified linear units and dropout (MLP). Employed molecular features were either:
- Coulomb matrices, which describe pairwise interactions between each atoms present in a conformer, based on the charge and coordinate of each atom,
- a variant of the bag-of-bonds (BoB) representation including the values of every bond length, bond angle, and dihedral angle present in a conformer.
In total, 16 models were built (4 ML algorithms × 2 types of molecular features × 2 molecular fragment types). They were evaluated using a 5-fold cross-validation scheme. In addition, key hyperparameters of each model were optimised using a random search strategy.
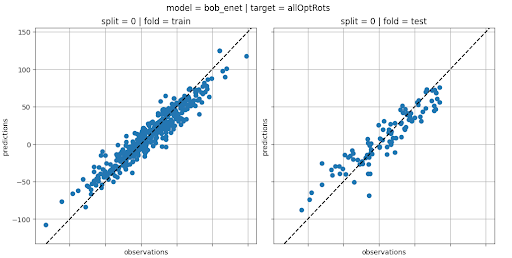
All supporting code was implemented using Python. Models were built using the Scikit Learn or Keras packages.
To interpret what molecular features were most influential on the predicted optical rotations, different strategies were used depending on the type of model. For the elastic net linear model, coefficient values were directly used. For nonlinear models (RF, ERT, MLP), the SHAP (SHapley Additive exPlanations) values approach was used.
This approach estimates the effect of each feature on each specific prediction for a given model, in an additive way. These estimates can then be aggregated to understand which molecular features influences optical rotation predictions the most (and how) for a particular model.
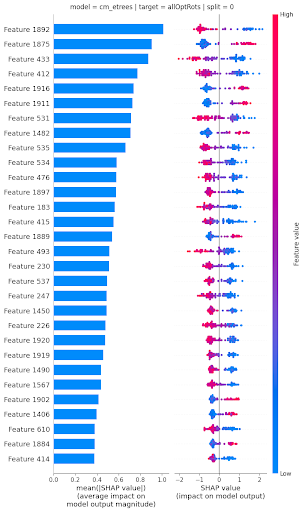
Looking at agreement between most influential features across models with the best predictive power allowed Benjamin to uncover that a small set of dihedral angles (angles between planes formed by triplets of atoms, with two atoms shared between the 2 triplets) influenced optical rotation values the most (see figure 1 at top of page). These findings have been included in a paper, currently under review.
Main outcomes
At the end of the consultancy, Maxime and Chris provided Benjamin with a full-featured ML pipeline tuned for his use case, allowing him to quickly explore a rich set of models to predict the optical properties based on molecular features. This pipeline was designed to be portable across platforms and leverage the computational power of an HPC platform for faster results.
The NeSI team also took great care in transferring knowledge to Benjamin so that he could apply these skills and approaches to future work. This included upskilling him in numerical Python software engineering, ML model implementation and ML pipeline design.
Researcher feedback
The collaboration with Maxime and Chris at NeSI has been highly beneficial for the progression of research into the optical properties of carbohydrate polymers. Their technical acumen in developing a machine learning pipeline has provided a robust framework for the analysis of multivariate and highly nonlinear data. I appreciate their methodical approach and the computational tools they were able to introduce to the project, which were essential in enabling the research and ensuring its continual progress thanks to their steadfast commitment.
– Dr Benjamin Westberry, Massey University
Do you want to bring your research to the next level? We can help. Send an email to support@nesi.org.nz to learn more about our Consultancy support.